Digital Documentation of COVID-19 Certificates (DDCC)
The Digital Documentation of COVID-19 Certificates (DDCC) published in August 2021 and similar guidance for test results (https://www.who.int/publications/i/item/WHO-2019-nCoV-Digital_certificates_diagnostic_test_results-2022.1) published in March 2022 serve as the reference for the associated FHIR Implementation Guide. This Implementation Guide (IG) acts as the computable representation of the core data elements in the published guidance, mapped to standard terminologies. These data models, in the form of FHIR logical models, and terminologies, in the form of FHIR value sets, are the basis for interoperability between the various credential formats. The IG provides FHIR Structure Maps and Concept Maps to define transformations between supported credential formats and the core data set, which allows business rules to execute against a common set of data elements.
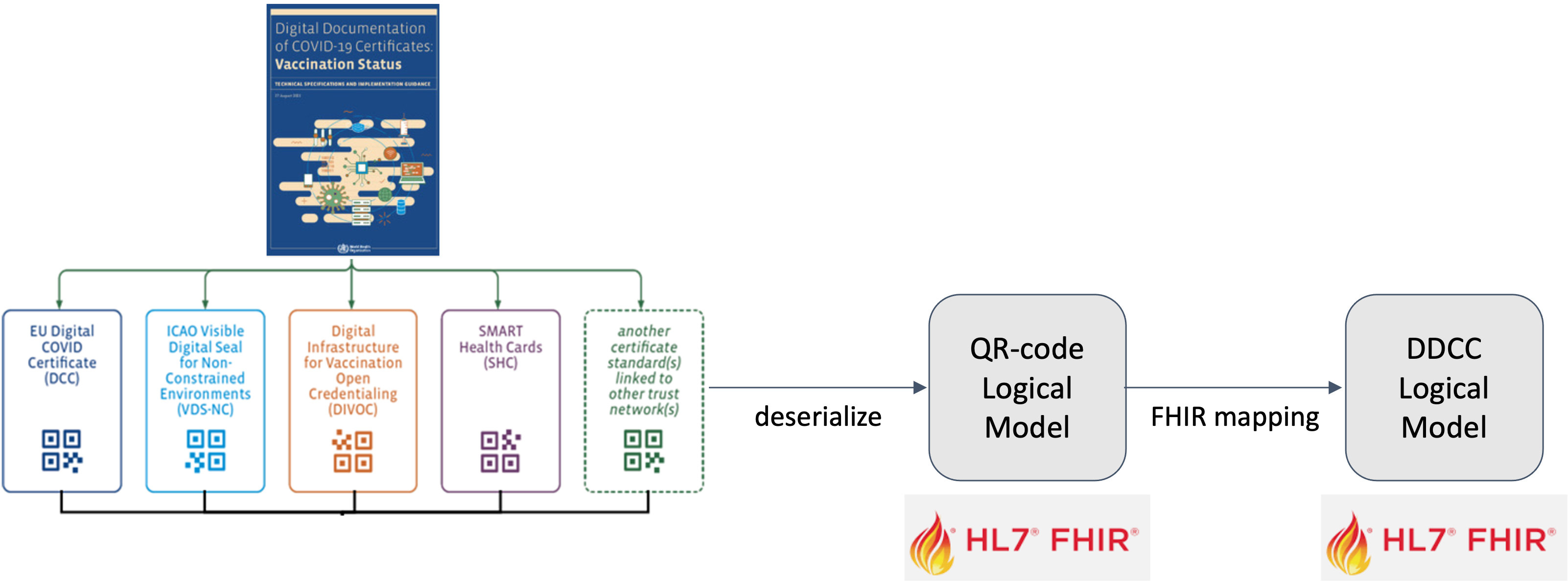
Strategy for mapping COVID certificate terminologies to the DDCC core data sets
Test result
The core data set (FHIR.WHO.DDCC\DDCC Core Data Set Logical Model for Test Result) has bindings to the following terminology:
Pathogen targeted (
pathogen):
ICD-11 MMS codes for COVID and COVID variants - FHIR.WHO.DDCC\WHO Agent Targeted (COVID-19)Type of test (
type):
ICD-11 Foundation Component entities for NAAT or rapid tests - FHIR.WHO.DDCC\WHO Type of Covid-19 TestSpecimen sample origin (
origin):
ICD-11 Foundation Component entities for the source of the specimen (e.g., nasopharyngeal swab) - FHIR.WHO.DDCC\WHO Speciman Sample Origin (COVID-19)Test result (
result):
ICD-11 MMS codes for "detected" or "not detected" - FHIR.WHO.DDCC\WHO Test Result (COVID-19)Test centre country (
country):
Three character country codes - Valueset-iso3166-1-3
| Cert. type | Element | Core data mapping | Approach |
|---|---|---|---|
| DIVOC | evidence.disease | pathogen | Map custom code system to ICD-11 |
| DIVOC | evidence.testType | type | Map custom code system to ICD-11 |
| DIVOC | evidence.sampleOrigin | origin | Map custom code system to ICD-11 |
| DIVOC | evidence.result | result | Map custom code system to ICD-11 |
| DIVOC | evidence.facility.address.addressCountry | country | ISO-3166 two or three character |
| EU DCC | -260.t.tg | pathogen | Map SNOMED CT to ICD-11 |
| EU DCC | -260.t.tt | type | Map generic LOINCs to ICD-11 |
| EU DCC | Not present | origin | n/a |
| EU DCC | -260.t.tr | result | Map SNOMED CT to ICD-11 |
| EU DCC | -260.t.to | country | ISO-3166 two character |
| ICAO | Not present | pathogen | n/a |
| ICAO | data.msg.tr.tc | type | Map custom code system to ICD-11 |
| ICAO | data.msg.tr.m | origin | Map custom code system to ICD-11 |
| ICAO | data.msg.tr.r | result | Map custom code system to ICD-11 |
| ICAO | data.msg.sp.ctr | country | Assuming ISO-3166 three character |
| SHC | Inferred from code | pathogen | Assume COVID-19 if LOINC listed here |
| SHC | code* | type | Map LOINC to ICD-11 |
| SHC | Not currently present, but will be bodySite* | origin | n/a |
| SHC | valueCodeableConcept.coding[0].code* | result | Map SNOMED CT to ICD-11 |
* Note that these elements are within the FHIR Bundle that is part of the SMART Health Card. The full path to this is vc.credentialSubject.fhirBundle.entry[].resource.
Proof of vaccination
The core data set (FHIR.WHO.DDCC\DDCC Core Data Set Logical Model - Vaccination Status - Proof of Vaccination) has bindings to the following terminology:
Vaccine (
vaccine):
ICD-11 MMS codes for both types of vaccine (e.g.,XM5DF6 COVID-19 vaccine, live attenuated virus) and specific vaccine products (e.g.,XM3DT5 COVID-19 Vaccine Moderna)Vaccine brand (
brand):
Not currently bound to a ValueSetCountry of vaccination (
country):
Three character country codes - Valueset-iso3166-1-3
Mapping approach
| Cert. type | Element | Core data mapping | Approach |
|---|---|---|---|
| DIVOC | evidence.icd11Code | vaccine | Presumably already ICD-11 MMS (see below) |
| DIVOC | evidence.facility.address.addressCountry | country | ISO-3166 two or three character |
| EU DCC | -260.1.v.mp | vaccine | Preferred binding is Union Register, but could also be ICD-11 or arbitrary strings. Only Union Register is considered in our mapping code. |
| EU DCC | -260.1.v.co | country | ISO-3166 two character |
| ICAO | data.msg.ve.des | vaccine | Already ICD-11 MMS |
| ICAO | data.msg.ve.vd[].ctr | country | Already ISO-3166 three character |
| SHC | vaccineCode.coding[0].code* | vaccine | ICD-11 MMS, CVX, or SNOMED CT; latter two mapped to ICD-11 MMS |
| SHC | Not present in computable form | country | n/a |
* Note that these elements are within the FHIR Bundle that is part of the SMART Health Card. The full path to this is vc.credentialSubject.fhirBundle.entry[].resource.
FHIR Resource naming conventions
ValueSets
- Lab test - pathogen (COVID-19):
name:LabTestPathogenCovid19{TerminologyName}ValueSetid:lab-test-pathogen-covid19-{terminology-name}title:Lab Test - Pathogen - COVID-19 - {Terminology Name}
- Lab test - type (COVID-19-specific):
name:LabTestTypeCovid19{TerminologyName}ValueSetid:lab-test-type-covid19-{terminology-name}title:Lab Test - Type - COVID-19 - {Terminology Name}
- Lab test - type (generic):
name:LabTestType{TerminologyName}ValueSetid:lab-test-type-{terminology-name}title:Lab Test - Type - {Terminology Name}
- Lab test - sample origin:
name:LabTestSampleOrigin{TerminologyName}ValueSetid:lab-test-sample-origin-{terminology-name}title:Lab Test - Sample Origin - {Terminology Name}
- Lab test - qualitative result:
name:LabTestQualitativeResult{TerminologyName}ValueSetid:lab-test-qual-result-{terminology-name}title:Lab Test - Qualitative Result - {Terminology Name}
- Vaccine (COVID-19):
name:VaccineCovid19{TerminologyName}ValueSetid:vaccine-covid19-{terminology-name}title:Vaccine - COVID-19 - {Terminology Name}
FSH filenames should be tx-vs-{id}.fsh. Each ValueSet should be in a separate file.
All ValueSet descriptions should describe where the codes in the value set came from and when the snapshot was taken. This should have enough information for someone to verify the contents of the value set or update it using the same approach.
The goal is that name should clearly identify both the resource type and contents when used in FSH within this IG, and the canonical URI (based on id) should not have any redundant information in it. The title should not include the resource type as this is implicit in how the resources are displayed in the IG.
Terminology names in ids should be shortened as much as possible while still being readable. For example, SNOEMD CT becomes snomed; ICD-11 becomes icd11.
"COVID-19" should appear in id/name/title when using a COVID-specific subset of a broader terminology system. For internally defined code systems (like those used by DIVOC and ICAO), "COVID-19" should not appear in case those specifications are expanded in the future (otherwise it would be necessary to create a 2nd value set, which would be problematic for terminology bindings in the logical model).
CodeSystems
Code systems are included to allow for expansion.
FSH filenames should be tx-cs-{alias}.fsh, like tx-cs-snomed.fsh. Each CodeSystem should be in a separate file. In cases where the IG already has a code system defined, the codes should be merged into the existing system rather than placed in a separate file.
ConceptMaps
In the naming scheme below:
$type=LabTestPathogenCovid19/lab-test-pathogen-covid19/Lab Test - Pathogen - COVID-19, etc.; for vaccines it isVaccineCovid19/vaccine-covid19/Vaccine - COVID-19$source= terminology name for the source terminology$target= terminology name for the target terminology
With these variables defined, ConceptMaps should be named as follows:
name:{$type}{$source}To{$target}ConceptMapid:{$type}-{$source}-to-{$target}title:{$type} - mapping {$source} to {$target}
For example, if:
$type=Vaccine - COVID-19(and variants)$source=CVX$target=ICD-11
...then we would have:
name:VaccineCovid19CVXToICD11ConceptMapid:vaccine-covid19-cvx-to-icd-11title:Vaccine - COVID-19 - mapping CVX to ICD-11- Filename:
tx-cm-vaccine-covid19-cvx-to-icd-11.fsh
Each ConceptMap should be in a separate file.
Source specifications
It can be difficult to find the "source of truth" specifications for the certificate formats that are mapped to the DDCC core data sets. The following are links to the best known documentation for each of the certificate formats listed above:
- DIVOC
- Elements are defined by JSON-LD. As of 2023-02-12, we were told by the DIVOC team that the most current JSON-LD context for vaccines is here and for test results is here.
- Note that the URIs for the DIVOC-specific JSON-LD contexts embedded in the certificates do not resolve.
- DIVOC's documentation is here, but this does not include element-level descriptions (e.g., terminology bindings at a per-element level).
- Terminology bindings at the element level must be inferred from more general documentation.
- The
v2JSON-LD context introduces theevidence.icd11Codeelement for vaccines, but there is no documentation on which codes this element is bound to or whether this is used in production. - There is no other computable representation of vaccine type/product.
- As of 2023-02-12, the public documentation does not include any information on test result certificates. The information we have was provided privately by the DIVOC team.
- Elements are defined by JSON-LD. As of 2023-02-12, we were told by the DIVOC team that the most current JSON-LD context for vaccines is here and for test results is here.
- EU DCC
- Starting place: Technical specifications for EU Digital COVID Certificates - Volumes 1-5.
- Volume 1: Formats and trust management references "Implementing Decision (EU) 2021/1073, Annex I".
- Implementing Decision (EU) 2021/1073, Annex I can be found here, along with other relevant Annexes.
- ICAO
- SMART Health Cards